- Explore MCP Servers
- pharmacology-mcp
Pharmacology Mcp
What is Pharmacology Mcp
pharmacology-mcp is an MCP (Model Context Protocol) server designed for the Guide to Pharmacology database, providing structured access to pharmacological data including targets, ligands, and interactions.
Use cases
Use cases for pharmacology-mcp include developing AI-driven applications for drug discovery, providing pharmacological insights in research projects, and enhancing educational tools with access to comprehensive pharmacological data.
How to use
To use pharmacology-mcp, connect to the server and utilize its API to query pharmacological data. Users can perform natural language queries to retrieve information about drug targets, ligands, interactions, and more.
Key features
Key features of pharmacology-mcp include structured access to authoritative pharmacological data sources, support for natural language queries, and type safety to ensure accurate data retrieval.
Where to use
pharmacology-mcp can be used in fields such as pharmacology, drug discovery, biomedical research, and AI applications that require access to specialized pharmacological knowledge.
Clients Supporting MCP
The following are the main client software that supports the Model Context Protocol. Click the link to visit the official website for more information.
Overview
What is Pharmacology Mcp
pharmacology-mcp is an MCP (Model Context Protocol) server designed for the Guide to Pharmacology database, providing structured access to pharmacological data including targets, ligands, and interactions.
Use cases
Use cases for pharmacology-mcp include developing AI-driven applications for drug discovery, providing pharmacological insights in research projects, and enhancing educational tools with access to comprehensive pharmacological data.
How to use
To use pharmacology-mcp, connect to the server and utilize its API to query pharmacological data. Users can perform natural language queries to retrieve information about drug targets, ligands, interactions, and more.
Key features
Key features of pharmacology-mcp include structured access to authoritative pharmacological data sources, support for natural language queries, and type safety to ensure accurate data retrieval.
Where to use
pharmacology-mcp can be used in fields such as pharmacology, drug discovery, biomedical research, and AI applications that require access to specialized pharmacological knowledge.
Clients Supporting MCP
The following are the main client software that supports the Model Context Protocol. Click the link to visit the official website for more information.
Content
pharmacology-mcp
MCP (Model Context Protocol) server for the Guide to PHARMACOLOGY database, providing access to pharmacological data including targets, ligands, and interactions.
This server implements the Model Context Protocol (MCP) for the Guide to PHARMACOLOGY, providing a standardized interface for accessing pharmacological data. MCP enables AI assistants and agents to access specialized pharmacological knowledge through structured interfaces to authoritative data sources.
The Guide to PHARMACOLOGY is an expert-curated database of drug targets and their ligands, providing comprehensive information about:
- Targets: Pharmacological targets (receptors, enzymes, ion channels, etc.)
- Ligands: Chemical compounds and drugs
- Interactions: Target-ligand interactions with affinity data
- Diseases: Disease associations
- Families: Target family classifications
If you want to understand more about what the Model Context Protocol is and how to use it more efficiently, you can take the DeepLearning AI Course or search for MCP videos on YouTube.
About MCP (Model Context Protocol)
MCP is a protocol that bridges the gap between AI systems and specialized domain knowledge. It enables:
- Structured Access: Direct connection to authoritative pharmacological data sources
- Natural Language Queries: Simplified interaction with specialized databases
- Type Safety: Strong typing and validation through FastMCP
- AI Integration: Seamless integration with AI assistants and agents
Quick Start
Installing uv
# Download and install uv
curl -LsSf https://astral.sh/uv/install.sh | sh
# Verify installation
uv --version
uvx --version
uvx is a very nice tool that can run a python package installing it if needed.
Running with uvx
You can run the pharmacology-mcp server directly using uvx without cloning the repository:
# Run the server in streamed http mode (default)
uvx pharmacology-mcp
Other uvx modes (STDIO, HTTP, SSE)
STDIO Mode (for MCP clients that require stdio, can be useful when you want to save files)
# Or explicitly specify stdio mode
uvx pharmacology-mcp stdio
HTTP Mode (Web Server)
# Run the server in streamable HTTP mode on default (3001) port
uvx pharmacology-mcp server
# Run on a specific port
uvx pharmacology-mcp server --port 8000
SSE Mode (Server-Sent Events)
# Run the server in SSE mode
uvx pharmacology-mcp sse
In cases when there are problems with uvx often they can be caused by cleaning uv cache:
uv cache clean
The HTTP mode will start a web server that you can access at http://localhost:3001/mcp (with documentation at http://localhost:3001/docs). The STDIO mode is designed for MCP clients that communicate via standard input/output, while SSE mode uses Server-Sent Events for real-time communication.
API Endpoints
The server provides REST API endpoints for:
- Targets: Pharmacological targets (receptors, enzymes, ion channels, etc.)
- Ligands: Chemical compounds and drugs
- Interactions: Target-ligand interactions with affinity data
- Diseases: Disease associations
- Families: Target family classifications
Example API Usage
import httpx
# Search for GPCR targets
async with httpx.AsyncClient() as client:
response = await client.post(
"http://localhost:8000/targets",
json={"type": "GPCR", "immuno": True}
)
targets = response.json()
# Get specific target information
async with httpx.AsyncClient() as client:
response = await client.get("http://localhost:8000/targets/1")
target = response.json()
# Search for approved drugs
async with httpx.AsyncClient() as client:
response = await client.post(
"http://localhost:8000/ligands",
json={"approved": True, "type": "Synthetic organic"}
)
ligands = response.json()
Usage Example
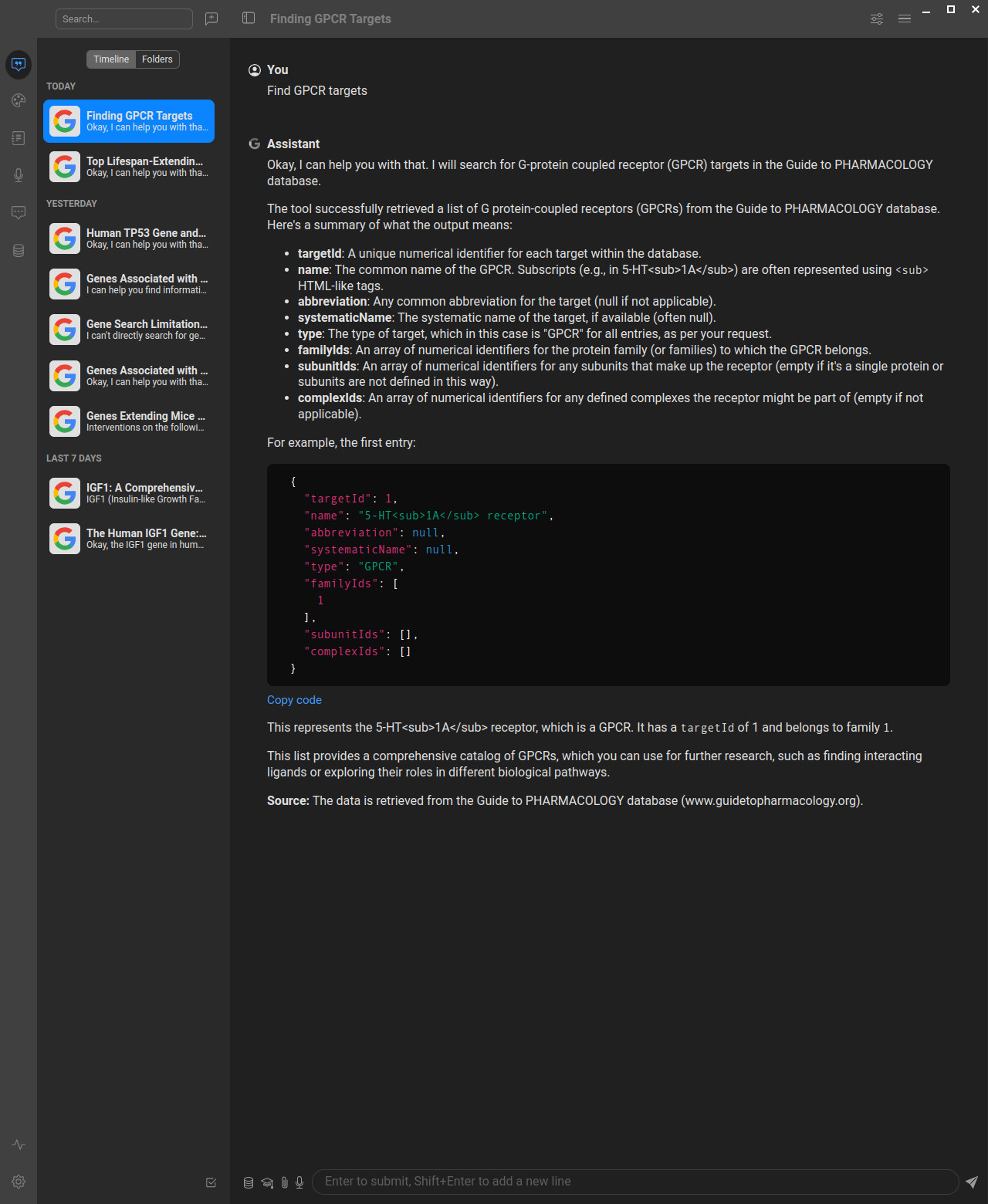
Local File Tools
The server also provides MCP tools for saving search results to files:
search_targets_to_file: Search targets and save resultssearch_ligands_to_file: Search ligands and save resultsget_target_interactions_to_file: Get target interactions and saveget_ligand_interactions_to_file: Get ligand interactions and save
Configuring your AI Client (Anthropic Claude Desktop, Cursor, Windsurf, etc.)
We provide preconfigured JSON files for different use cases:
- For STDIO mode (recommended): Use
mcp-config-stdio.json - For HTTP mode: Use
mcp-config.json - For local development: Use
mcp-config-stdio-debug.json
Inspecting Pharmacology MCP server
Using MCP Inspector to explore server capabilities
If you want to inspect the methods provided by the MCP server, use npx (you may need to install nodejs and npm):
For STDIO mode with uvx:
npx @modelcontextprotocol/inspector --config mcp-config-stdio.json --server pharmacology-mcp
For HTTP mode (ensure server is running first):
npx @modelcontextprotocol/inspector --config mcp-config.json --server pharmacology-mcp
For local development:
npx @modelcontextprotocol/inspector --config mcp-config-stdio-debug.json --server pharmacology-mcp
You can also run the inspector manually and configure it through the interface:
npx @modelcontextprotocol/inspector
After that you can explore the tools and resources with MCP Inspector at http://127.0.0.1:6274
Integration with AI Systems
Simply point your AI client (like Cursor, Windsurf, ClaudeDesktop, VS Code with Copilot, or others) to use the appropriate configuration file from the repository.
Repository setup
# Clone the repository
git clone https://github.com/antonkulaga/pharmacology-mcp.git
cd pharmacology-mcp
uv sync
Running the MCP Server
If you already cloned the repo you can run the server with uv:
# Start the MCP server locally (HTTP mode)
uv run server
# Or start in STDIO mode
uv run stdio
# Or start in SSE mode
uv run sse
Testing & Verification
Run tests for the MCP server:
uv run pytest -vvv -s
You can use MCP inspector with locally built MCP server same way as with uvx.
Note: Using the MCP Inspector is optional. Most MCP clients (like Cursor, Windsurf, etc.) will automatically display the available tools from this server once configured. However, the Inspector can be useful for detailed testing and exploration.
If you choose to use the Inspector via npx, ensure you have Node.js and npm installed. Using nvm (Node Version Manager) is recommended for managing Node.js versions.
API Documentation
When the server is running, you can access:
- Interactive API docs: http://localhost:8000/docs
- ReDoc documentation: http://localhost:8000/redoc
- OpenAPI schema: http://localhost:8000/openapi.json
Data Source
This server provides access to data from the Guide to PHARMACOLOGY, an expert-curated database of drug targets and their ligands.
License
This project is licensed under the MIT License.
- Database: Open Data Commons Open Database License (ODbL)
- Contents: Creative Commons Attribution-ShareAlike 4.0 International License
Citations
If you use this server in your research, please cite:
Armstrong JF, Faccenda E, Harding SD, Pawson AJ, Southan C, Sharman JL, Campo B, Cavanagh DR, Alexander SPH, Davenport AP, Spedding M, Davies JA; NC-IUPHAR. (2020) The IUPHAR/BPS Guide to PHARMACOLOGY in 2020: extending immunopharmacology content and introducing the IUPHAR/MMV Guide to MALARIA PHARMACOLOGY. Nucleic Acids Research 48: D1006-D1021.
Contributing
Contributions are welcome! Please feel free to submit a Pull Request.
Acknowledgments
- Guide to PHARMACOLOGY for the comprehensive pharmacological data
- Model Context Protocol for the protocol specification
- FastMCP for the MCP server framework
This project is part of the Longevity Genie organization, which develops open-source AI assistants and libraries for health, genetics, and longevity research.
Dev Tools Supporting MCP
The following are the main code editors that support the Model Context Protocol. Click the link to visit the official website for more information.












